category
药物发现是一个复杂、耗时的过程,需要研究人员浏览大量的科学文献、临床试验数据和分子数据库。基因泰克和阿斯利康等生命科学客户正在使用人工智能代理和其他生成性人工智能工具来提高科学发现的速度。这些组织的建设者已经在使用Amazon Bedrock的完全托管功能,为各种用例快速部署特定领域的工作流程,从早期药物靶点识别到医疗保健提供者参与。
然而,更复杂的用例可能会从使用开源Strands Agents SDK中受益。Strands Agents采用模型驱动的方法来开发和运行AI代理。它适用于大多数模型提供者,包括自定义和内部大型语言模型(LLM)网关,并且可以在托管Python应用程序的地方部署代理。
在这篇文章中,我们将演示如何使用Strands Agents和Amazon Bedrock创建一个强大的药物发现研究助理。该AI助手可以使用模型上下文协议(MCP)同时搜索多个科学数据库,综合其发现,并生成有关药物靶点、疾病机制和治疗领域的综合报告。此助手可作为开源医疗保健和生命科学代理工具包中的示例,供您使用和调整。
解决方案概述
该解决方案使用Strands Agent将高性能基础模型(FM)与arXiv、PubMed和ChEMBL等常见生命科学数据源连接起来。它演示了如何快速创建MCP服务器来查询数据并在对话界面中查看结果。
协同工作的小型、专注的人工智能代理通常可以产生比单个、单片代理更好的结果。此解决方案使用一组子代理,每个子代理都有自己的FM、说明和工具。以下流程图显示 how the orchestrator agent (shown in orange) handles user queries and routes them to sub-agents for either information retrieval (green) or planning, synthesis, and report generation (purple).

This post focuses on building with Strands Agents in your local development environment. Refer to the Strands Agents documentation to deploy production agents on AWS Lambda, AWS Fargate,
Amazon Elastic Kubernetes Service (Amazon EKS), or Amazon Elastic Compute Cloud (Amazon EC2).
In the following sections, we show how to create the research assistant in Strands Agents by defining an FM, MCP tools, and sub-agents.
Prerequisites
This solution requires Python 3.10+, strands-agents, and several additional Python packages. We strongly recommend using a virtual environment like venv or uv to manage these dependencies.
Complete the following steps to deploy the solution to your local environment:
- Clone the code repository from GitHub.
- Install the required Python dependencies with
pip install -r requirements.txt. - Configure your AWS credentials by setting them as environment variables, adding them to a credentials file, or following another supported process.
- Save your Tavily API key to a .env file in the following format:
TAVILY_API_KEY="YOUR_API_KEY".
You also need access to the following Amazon Bedrock FMs in your AWS account:
- Anthropic’s Claude 3.7 Sonnet
- Anthropic’s Claude 3.5 Sonnet
- Anthropic’s Claude 3.5 Haiku
Define the foundation model
We start by defining a connection to an FM in Amazon Bedrock using the Strands Agents BedrockModel class. We use Anthropic’s Claude 3.7 Sonnet as the default model. See the following code:
Define MCP tools
MCP provides a standard for how AI applications interact with their external environments. Thousands of MCP servers already exist, including those for life science tools and datasets. This solution provides
example MCP servers for:
- arXiv – Open-access repository of scholarly articles
- PubMed – Peer-reviewed citations for biomedical literature
- ChEMBL – Curated database of bioactive molecules with drug-like properties
- ClinicalTrials.gov – US government database of clinical research studies
- Tavily Web Search – API to find recent news and other content from the public internet
Strands Agents streamlines the definition of MCP clients for our agent. In this example, you connect to each tool using standard I/O. However, Strands Agents also supports
remote MCP servers with Streamable-HTTP Events transport. See the following code:
Define specialized sub-agents
The planning agent looks at user questions and creates a plan for which sub-agents and tools to use:
Similarly, the synthesis agent integrates findings from multiple sources into a single, comprehensive report:
Define the orchestration agent
We also define an orchestration agent to coordinate the entire research workflow. This agent uses the SlidingWindowConversationManager class from Strands
Agents to store the last 10 messages in the conversation. See the following code:
Example use case: Explore recent breast cancer research
To test out the new assistant, launch the chat interface by running streamlit run application/app.py and opening the local URL (typically http://localhost:8501) in your web browser. The following
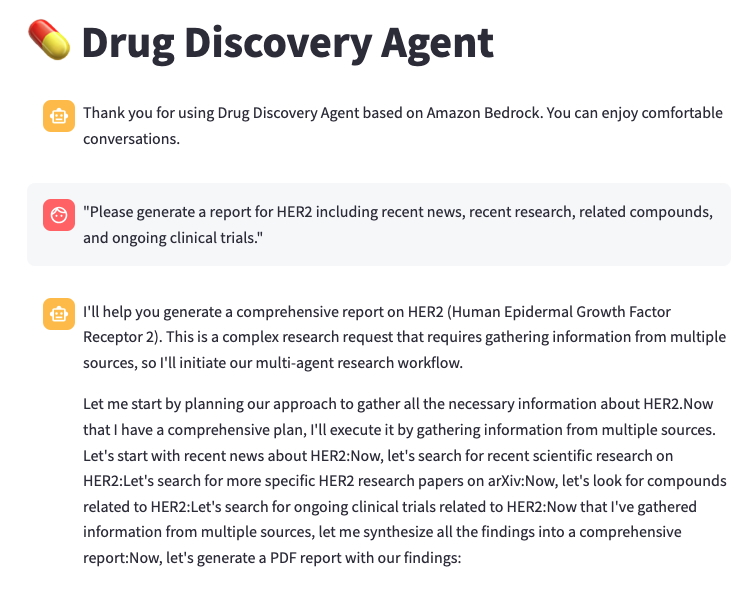
screenshot shows a typical conversation with the research agent. In this example, we ask the assistant, “Please generate a report for HER2 including recent news, recent research,
related compounds, and ongoing clinical trials.” The assistant first develops a comprehensive research plan using the various tools at its disposal. It decides to start with a web search
for recent news about HER2, as well as scientific articles on PubMed and arXiv. It also looks at HER2-related compounds in ChEMBL and ongoing clinical trials. It synthesizes these
results into a single report and generates an output file of its findings, including citations.
The following is an excerpt of a generated report:
Notably, you don’t have to define a step-by-step process to accomplish this task. By providing the assistant with a well-documented list of tools, it can decide which to use and in what order.
Clean up
If you followed this example on your local computer, you will not create new resources in your AWS account that you need to clean up. If you deployed the research assistant using one of those services,
refer to the relevant service documentation for cleanup instructions.
Conclusion
In this post, we showed how Strands Agents streamlines the creation of powerful, domain-specific AI assistants. We encourage you to try this solution with your own research questions
and extend it with new scientific tools. The combination of Strands Agents’s orchestration capabilities, streaming responses, and flexible configuration with the powerful language
models of Amazon Bedrock creates a new paradigm for AI-assisted research. As the volume of scientific information continues to grow exponentially, frameworks like
Strands Agents will become essential tools for drug discovery.
To learn more about building intelligent agents with Strands Agents, refer to Introducing Strands Agents, an Open Source AI Agents SDK, Strands Agents SDK,
and the GitHub repository. You can also find more sample agents for healthcare and life sciences built on Amazon Bedrock.
For more information about implementing AI-powered solutions for drug discovery on AWS, visit us at AWS for Life Sciences.
- 登录 发表评论
- 11 次浏览